Specifications:
| Application | Next Generation Sequencing, NGS Library Preparation |
| Storage Temperature | -20°C |
| Product Type | NGS library Prep kits |
| Product Brand | Hologic Diagenode |
| Product Grade | Molecular Biology |
ATAC-seq, Assay for Transposase-Accessible Chromatin, followed by next generation sequencing, is a key technology for genome-wide mapping of accessible chromatin. The technology is based on the use of the transposase Tn5 which cuts exposed open chromatin and simultaneously ligates adapters for subsequent amplification and sequencing. ATAC-seq methods allow you to:
- Gain insight into gene regulation and understand open chromatin signatures
- Determine nucleosome positions at single nucleotide resolution
- Uncover transcription factor (TF) occupancy
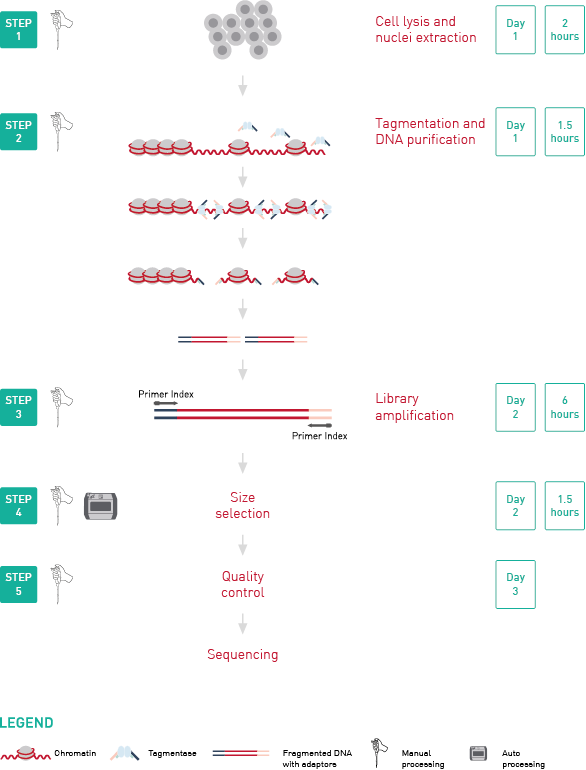
Diagenode’s ATAC-seq kit is based on a highly validated protocol optimized for 50,000 cells per reaction. The kit includes the reagents for cell lysis and nuclei extraction, tagmentation and DNA purification as well as for library amplification. The primer indexes for multiplexing are not included in the kit and must be purchased separately.
ATAC-seq kit features:
- Cell requirement: 50,000 cells / rxn
- Robust protocol with high reproducibility between replicates and repetitive experiments
- Easy and efficient DNA capture after the tagmentation reaction using Diagenode`s MicroChIP DiaPure columns (included)
- Additional qPCR step to determine the number of cycles needed for library amplification:
- Avoids over-amplification
- Allows adaptation/flexibility for more challenging samples to succeed with library prep.
- Gives early indication if the experiment does not work (no qPCR amplification)
ATAC-seq, Assay for Transposase-Accessible Chromatin, followed by next generation sequencing, is a key technology to easily identify the open regions of the chromatin. The protocol consists of 3 steps: nuclei preparation, tagmentation and library amplification. First, the cells undergo the lysis, ending with the crude nuclei. Then, the nuclei are incubated with a tagmentase (Tn5 transposase), which cuts the genomic regions associated with open chromatin and inserts the sequencing adaptors. Finally, the generated libraries are amplified and can be used for sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only, giving a map of the chromatin status in the whole genome of the sample.
Example of results
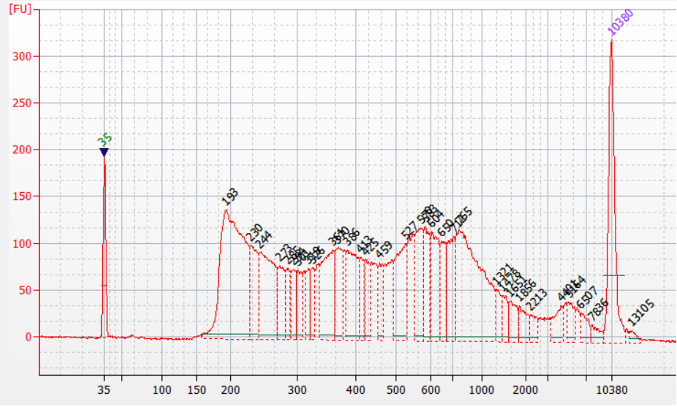
Figure 1.Representative Bioanalyzer profile of an ATAC-seq library prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.
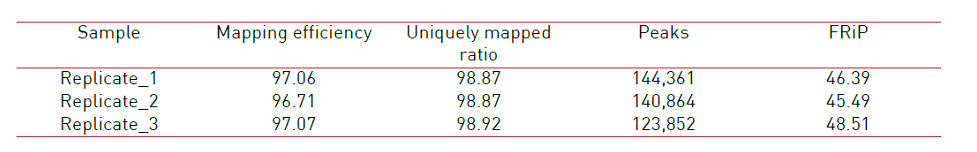
Figure 2. Main ATAC-seq alignment and peak calling statistics of 3 replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells. (Mapping efficiency: Percentage of non-mitochondrial reads that mapped to the reference genome. Uniquely mapped ratio: Proportion of mapped reads that map to only one location on the reference genome (hg19). Peaks: Number of peaks (open chromatin regions) identified by MACS2 for each sample. FRiP - Fraction of reads in peaks: Percentage of reads in peaks, with respect to the number of uniquely mapped reads. Sequencing was realized in paired-end mode 50 base pairs (PE50) on an Illumina NovaSeq6000.)
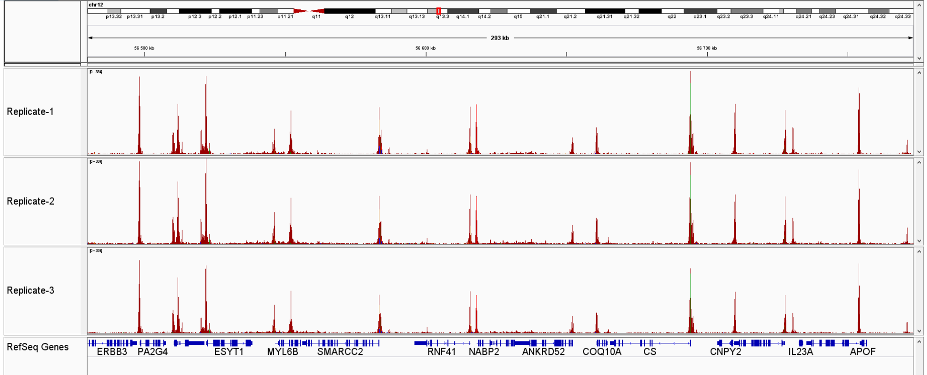
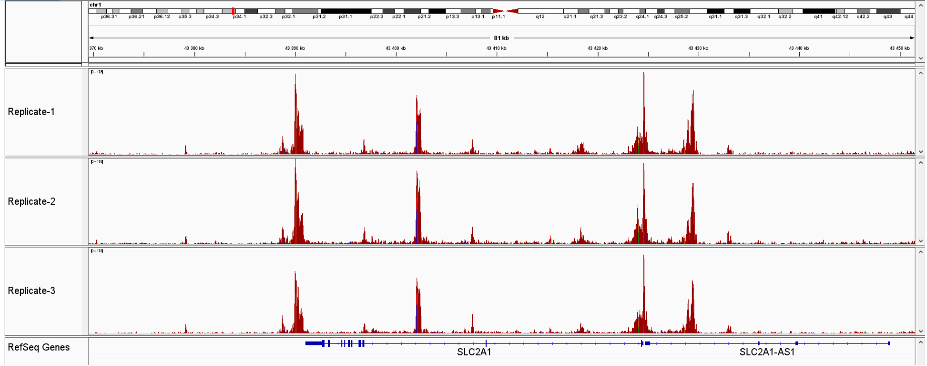
Figure 3 Sequencing profiles of ATAC-seq library (3 replicates) prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.
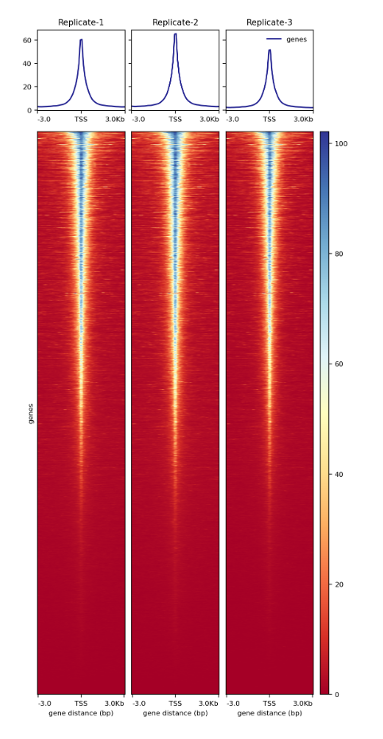
Figure 4.
Heatmap around TSS of three ATAC-seq replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.
- Reactions: 8 Rxns 24 Rxns




 0
0
