Diagenode iDeal ChIP-seq kit for Transcription Factors
Catalog No :
CAS Number :
Brand :
In Stock
Specifications:
| Application | Next Generation Sequencing, NGS Library Preparation |
| Storage Temperature | -20°C |
| Product Type | NGS library Prep kits |
| Product Brand | Hologic Diagenode |
| Product Grade | Molecular Biology |
Diagenode’s iDeal ChIP-seq Kit for Transcription Factors is a highly validated solution for robust transcription factor and other non-histone proteins ChIP-seq results and contains everything you need for start-to-finish ChIP prior to Next-Generation Sequencing. This complete solution contains all buffers and reagents for cell lysis, chromatin shearing, immunoprecipitation, and DNA purification. In addition, unlike competing solutions, the kit contains positive and negative control antibodies (CTCF and IgG, respectively) as well as positive and negative control PCR primers pairs (H19 and Myoglobin exon 2, respectively) for your convenience and a guarantee of optimal results.
The iDeal ChIP-seq kit for Transcription Factors is compatible for cells or tissues:
| Amount per IP | |
| Cells | 4,000,000 |
| Tissues | 30 mg |
The iDeal ChIP-seq kit is the only kit on the market validated for major sequencing systems.
- Highly optimized protocol for ChIP-seq from cells and tissues
- Validated for ChIP-seq with multiple transcription factors and non-histone targets
- Most complete kit available (covers all steps, including the control antibodies and primers)
- Magnetic beads make ChIP easy, fast and more reproducible
- Combination with Diagenode ChIP-seq antibodies provides high yields with excellent specificity and sensitivity
- Purified DNA suitable for any downstream application
- Easy-to-follow protocol
ChIP-seq on cells
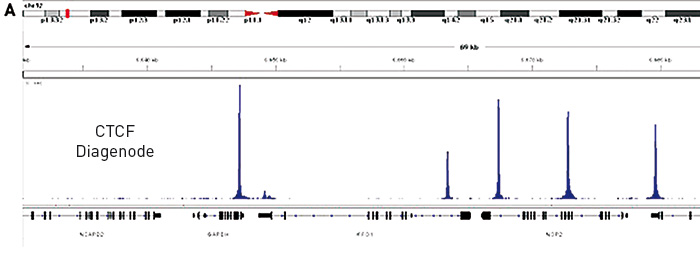
Figure 1. (A) Chromatin Immunoprecipitation has been performed using chromatin from HeLa cells, the iDeal ChIP-seq kit for Transcription Factors and the Diagenode ChIP-seq-grade CTCF antibody. The IP'd DNA was subsequently analysed on an Illumina® HiSeq. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. This figure shows the peak distribution in a region surrounding the GAPDH positive control gene.
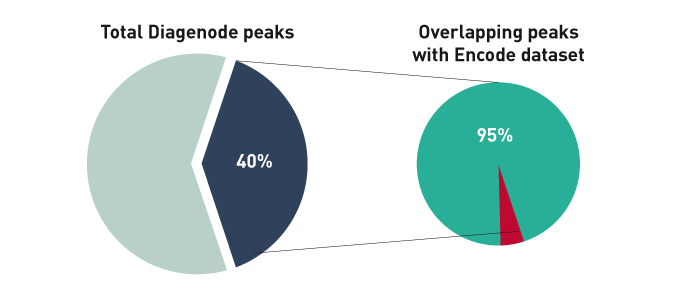
Figure 1B. The ChIP-seq dataset from this experiment has been compared with a reference dataset from the Broad Institute. We observed a perfect match between the top 40% of Diagenode peaks and the reference dataset. Based on the NIH Encode project criterion, ChIP-seq results are considered reproducible between an original and reproduced dataset if the top 40% of peaks have at least an 80% overlap ratio with the compared dataset.
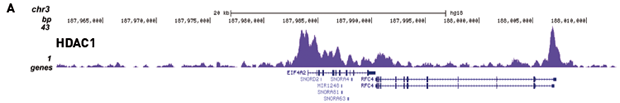
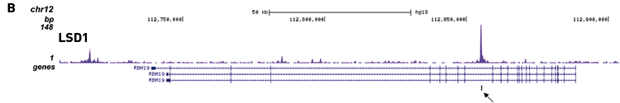

Figure 2. Chromatin Immunoprecipitation has been performed using chromatin from HeLa cells, the iDeal ChIP-seq kit for Transcription Factors and the Diagenode ChIP-seq-grade HDAC1 (A), LSD1 (B) and p53 antibody (C). The IP'd DNA was subsequently analysed on an Illumina® Genome Analyzer. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. This figure shows the peak distribution in regions of chromosome 3 (A), chromosome 12 (B) and chromosome 6 (C) respectively.
ChIP-seq on tissue
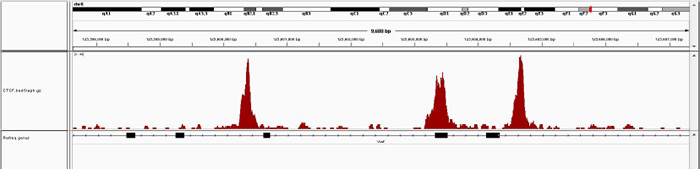
Figure 3A. Chromatin Immunoprecipitation has been performed using chromatin from mouse liver tissue, the iDeal ChIP-seq kit for Transcription Factors and the Diagenode ChIP-seq-grade CTCF antibody. The IP'd DNA was subsequently analysed on an Illumina® HiSeq. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. This figure shows the peak distribution in a region surrounding the Vwf positive control gene.
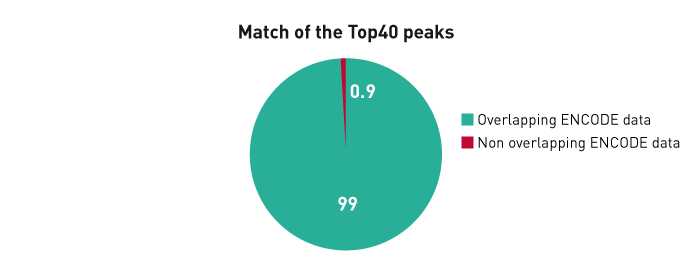
Figure 3B. The ChIP-seq dataset from this experiment has been compared with a reference dataset from the Broad Institute. We observed a perfect match between the top 40% of Diagenode peaks and the reference dataset. Based on the NIH Encode project criterion, ChIP-seq results are considered reproducible between an original and reproduced dataset if the top 40% of peaks have at least an 80% overlap ratio with the compared dataset.
The iDeal ChIP-seq Kit for Transcription Factors is compatible with a broad variety of cell lines, tissues and species - some examples are shown below. Other species / cell lines / tissues can be used with this kit.
Cell lines:
Human: A549, A673, BT-549, CD4 T, HCC1806, HeLa, HepG2, HFF, HK-GFP-MR, ILC, K562, KYSE-180, LapC4, M14, MCF7, MDA-MB-231, MDA-MB-436, RDES, SKNO1, VCaP, U2-OS, ZR-75-1
Mouse: ESC, NPCs, BZ, GT1-7, acinar cells, HSPCs, Th2 cells, keratinocytes
Cattle: pbMEC, MAC-T
Other cell lines / species: compatible, not tested
Tissues:
Mouse: kidney, heart, brain, iris, liver, limbs from E10.5 embryos
Horse: liver, brain, heart, lung, skeletal muscle, lamina, ovary
Other tissues: compatible, not tested
ChIP on yeast
The iDeal ChIP-seq kit for TF is compatible with yeast samples. Check out the Application Note presenting an optimized detailed protocol for ChIP on yeast.
- Reactions: 100 rxn 10 rxn 24 Rxns




 0
0
